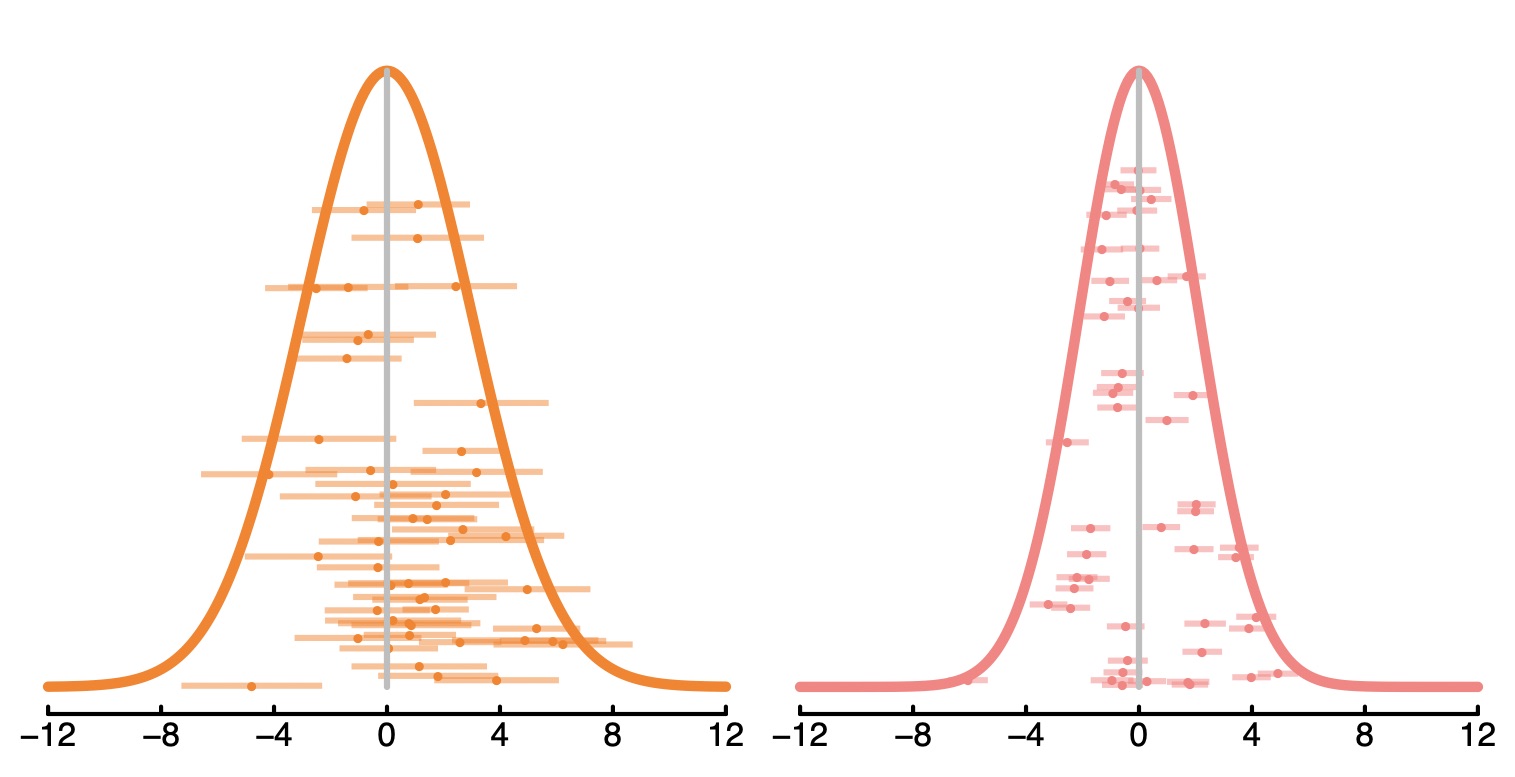
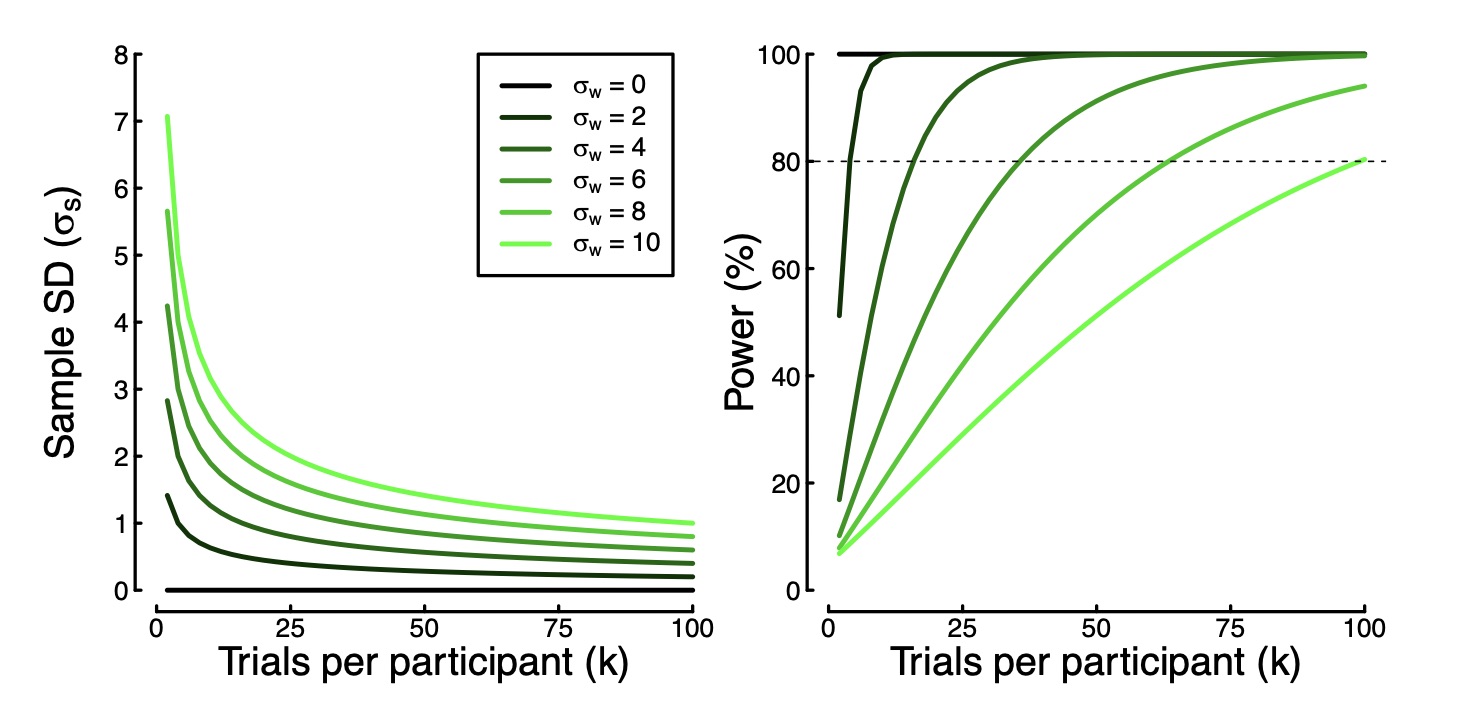
A few years back I was reading a stats paper. It mentioned somewhere that the entire paper was computationally reproducible. In other words, there was a single script sitting on github that could generate the whole paper from scratch. It will do all of the simulations, plot all of the figures, and produce a pdf of the manuscript. This is a really important step forward for transparency – the reader can see exactly what analyses have taken place, and even change them to see how this affects the outcome.
I was captivated by this idea, and I started thinking about how to apply it to empirical work. Stats papers are one thing, as they can often be based on simulations that don’t require any external data. But for empirical studies, the ‘holy grail’ would be to download all of the raw data and do all analyses entirely from scratch, all automatically. This turns out to be quite a bit of work! However we now have some published examples of fully computationally reproducible studies (see links below). I thought it was worth putting together some notes about our experiences to hopefully guide others in doing the same thing.
Software
A huge advance that helps make computational reproducibility possible is the existence of markdown documents. The basic idea is that you can create a single document that combines normal text, images, equations, and sections of computer code. When the script is executed, the code gets run, and an output document is produced containing the results. The output can be in any arbitrary format, including html, pdf and word processor document formats.
There are also many different flavours of markdown. So far I have mostly used Rmarkdown, which is well integrated with the RStudio application. However there are similar offerings for Python (e.g. Jupyter and PyPI), Matlab (live scripts), and from Google (Colab). It’s also possible to execute code snippets from other languages within an R markdown document if required.
Markdown does a lot of the heavy lifting in combining the code and text underlying a paper. It also uses LaTeX as an intermediate step, e.g. when producing a PDF file. That makes it possible to import the LaTeX script into another editor like Overleaf, which I’ve found is useful for applying journal style templates. It also means that equations can be typeset using LaTeX, so they look how they’re supposed to look.
Storage for code and data
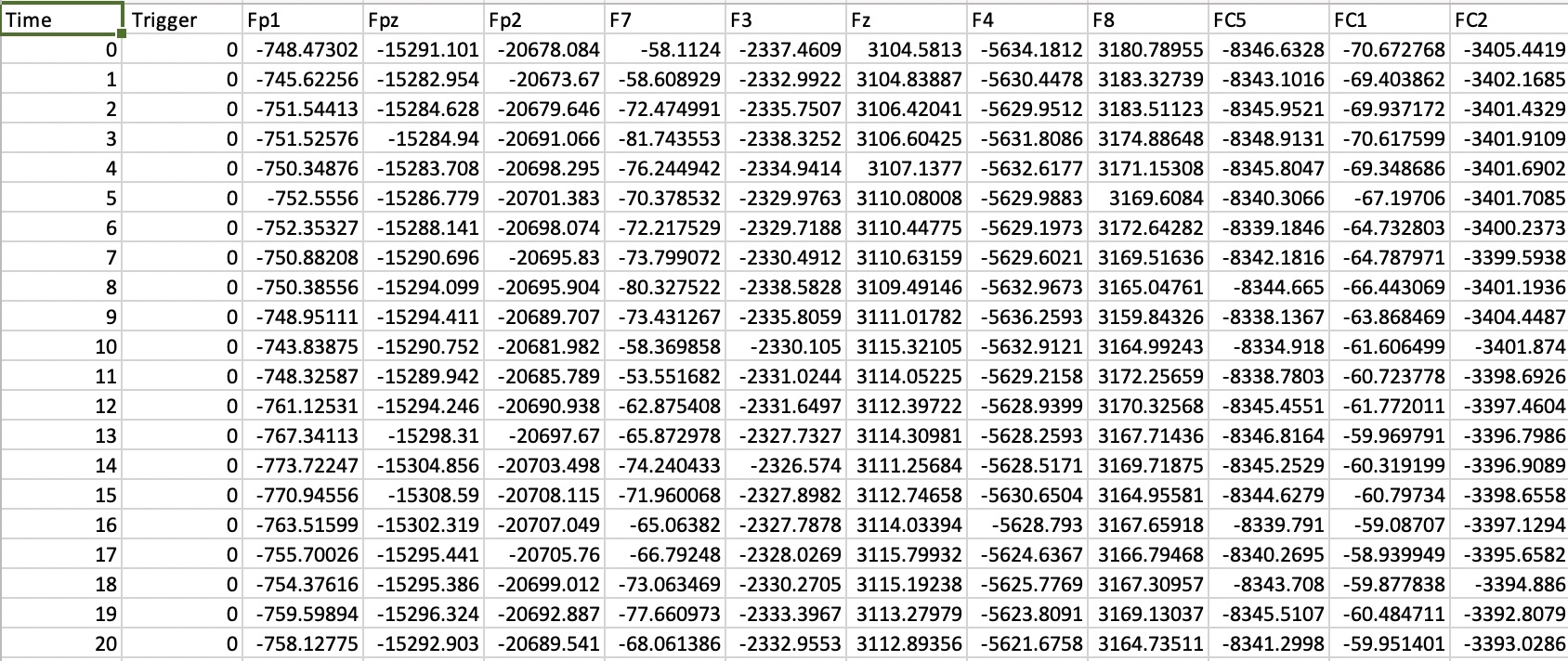
One important piece of the puzzle in realising reproducible empirical papers is how to automatically download the raw data. Github is great for storing code, but it isn’t really cut out for data storage, and has some quite harsh restrictive capacity limits for each project. It is viable for small data files, such as a spreadsheet containing data from a psychophysics experiment, or a questionnaire. But data files from studies using EEG, MRI, MEG, psychophysiology, eyetracking and other methods are often much larger, so including them in a github repo isn’t an option.
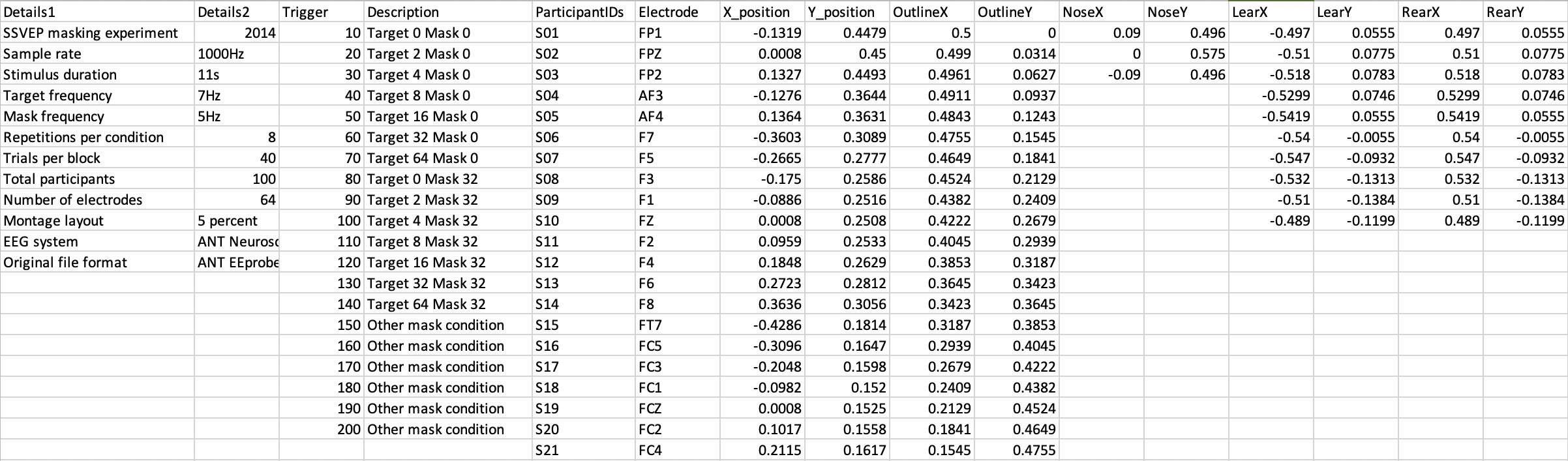
A solution I’ve found extremely useful is the osfr package that allows easy access to repositories on the OSF website. With a few lines of code it’s possible to automatically download files from any public repository on the site without needing any login credentials or access tokens (you do need an access token linked to your account in order to upload files). I think this makes life much easier for producing a fully automated script, because you don’t need to instruct the user to download a separate data file (or whole set of files) and put it in a specific location on their computer. Instead the script can grab whatever it needs and put it where it wants it, without any burden on the end user. I’m sure there are similar methods for other repositories, but this one seemed particularly frictionless. It’s also easy to link an OSF project to a github repo, so you can see the code in the same place as everything else.
A word of caution though – I have found that recently there is an occasional bug in indexing the files in some OSF repositories. Sometimes files are missed from a listing, though it’s not clear to me why this happens or what to do about it (see also this bug report: https://github.com/ropensci/osfr/issues/142). In the worst case, it might be necessary to manually download some of the data files, which isn’t exactly ideal. Hopefully this issue will be resolved by a package update.
Option flags to save time
Although the ultimate goal of computational reproducibility is that all of the analysis is done from scratch on the raw data, it’s not necessarily the case that everyone wants to do this all the time. In particular some analyses take several hours, and might need many gigabytes of storage space. I’ve found it useful to include a flag at the start of the script that specifies the level of analysis required. The current iteration of this idea has four levels:
3: Download all raw data and do all analysis from scratch
2: Do statistics, modelling, bootstrapping etc., but using processed data files that are smaller in size
1: Generate figures using processed data and the results of modelling (and other processing)
0: Don’t execute any analysis code, just generate a pdf of the manuscript with pre-generated figures
Using ‘if’ statements throughout the code lets these flags select which sections get executed. The flags turn out to also be extremely useful when writing the paper itself. For example you can avoid having to run analyses taking several hours when all you want to do is edit the text of the manuscript. It does need a bit of thought to make sure that processed data files can also be downloaded from the OSF repo if necessary, rather than forcing the user to go from the raw data if they don’t have the time (or the storage space). But overall this makes the code much easier to work with, and hopefully more straightforward for others.
Barriers to reproducibility
As with anything there are some limitations, for example some analyses use specialist software, and toolboxes that need user input, that cannot be straightforwardly integrated into a markdown pipeline. There are also restrictions on which data can be safely shared, for example structural MRI scans might plausibly be used to de-anonymise data if not processed appropriately before sharing. And I’m aware that writing bespoke analysis code is not everyone’s cup of tea, and is perhaps particularly well suited to certain data types (such as psychophysical or behavioural data, and SSVEP data). These caveats aside though, I think reproducibility is an important step towards a more open scientific landscape, and a worthy goal to aspire to.
Future plans
I was recently awarded some money from my University’s Enhancing Research Culture fund to pilot a project on computational reproducibility. The plan is to make ten studies computationally reproducible, using some of the methods described above, and also to push the envelope a bit in terms of technical skills. One particular goal is to start preserving the specific programming environment used, including all package versions (see this great paper for more details). Just like Nick Fury I’ve put a team together (of superhero PhD students) to work together on what I think will be an exciting project!
Examples of computationally reproducible papers:
Baker, D.H. (2021). Statistical analysis of periodic data in neuroscience. Neurons, Behavior, Data Analysis and Theory, 5(3): 1-18, [DOI] [code].
Baker, D.H., Vilidaite, G. & Wade, A.R. (2021). Steady-state measures of visual suppression. PLoS Computational Biology, 17(10): e1009507, [DOI] [code].
Baker, D.H. et al. (2023). Temporal dynamics of normalization reweighting. BioRxiv preprint, [DOI] [code].
Segala, F.G., Bruno, A., Wade, A.R. & Baker, D.H. (2023). Different rules for binocular combination of luminance in cortical and subcortical pathways. BioRxiv preprint, [DOI] [code].

 Posted by bakerdh
Posted by bakerdh